Hi all,
I have been tackling this problem for a few weeks now, to no avail. I’ve read up on similar orientation posts here and on the older MRTrix 2 FAQ and still have yet to solve the issue. So, I’m posting here hoping some of the experts can help.
I have rat DWI from a Bruker machine, which is converted to ANALYZE using the pconv tool, then converted to NIFTi via an in-house script which multiplies the voxel size by 10, applies orientation labels, generates sform/qform, and reorient the data via fslswapdim. With the reorientation, of course we’d need to double and triple check our bvecs and bvals, and have settled on a working bvec which gives the correct orientations using known anatomical landmarks. All of this, so far, is done through FSL, and is our current working model.
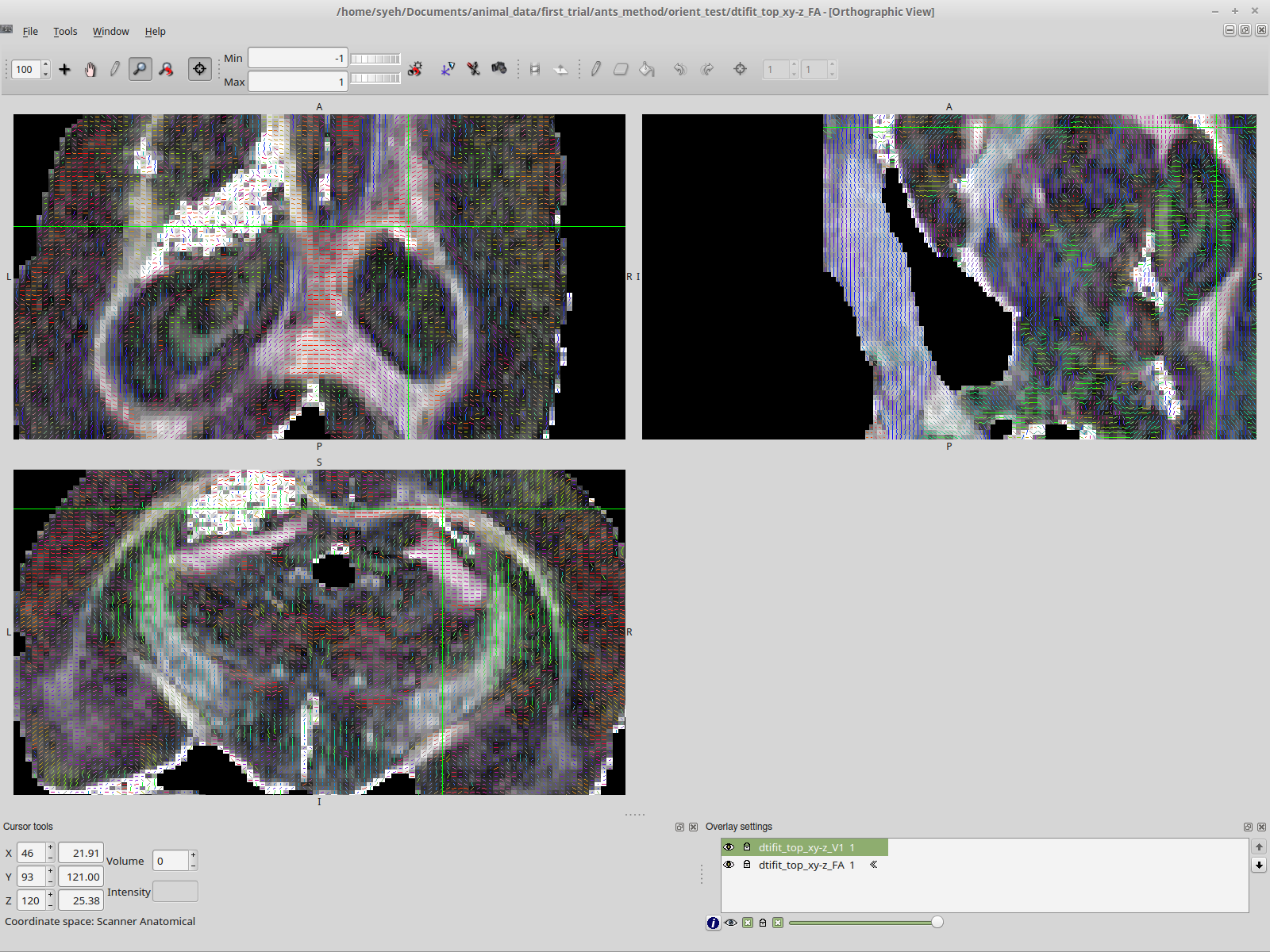
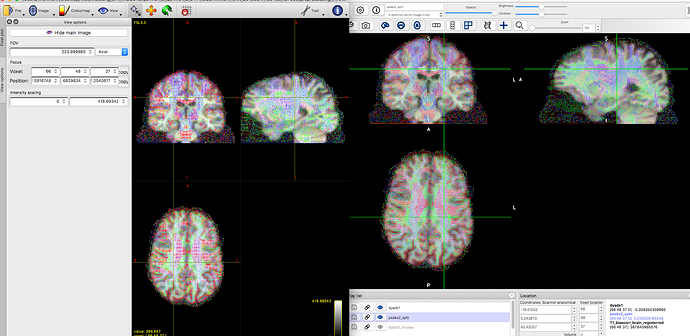
Vectors generated using FSL’s DTIFIT. Note the blue nerve bundle running anterior-posteriorly.Now, we are attempting to replicate the result in MRTrix, and almost everything is the same, except for the vector colors.
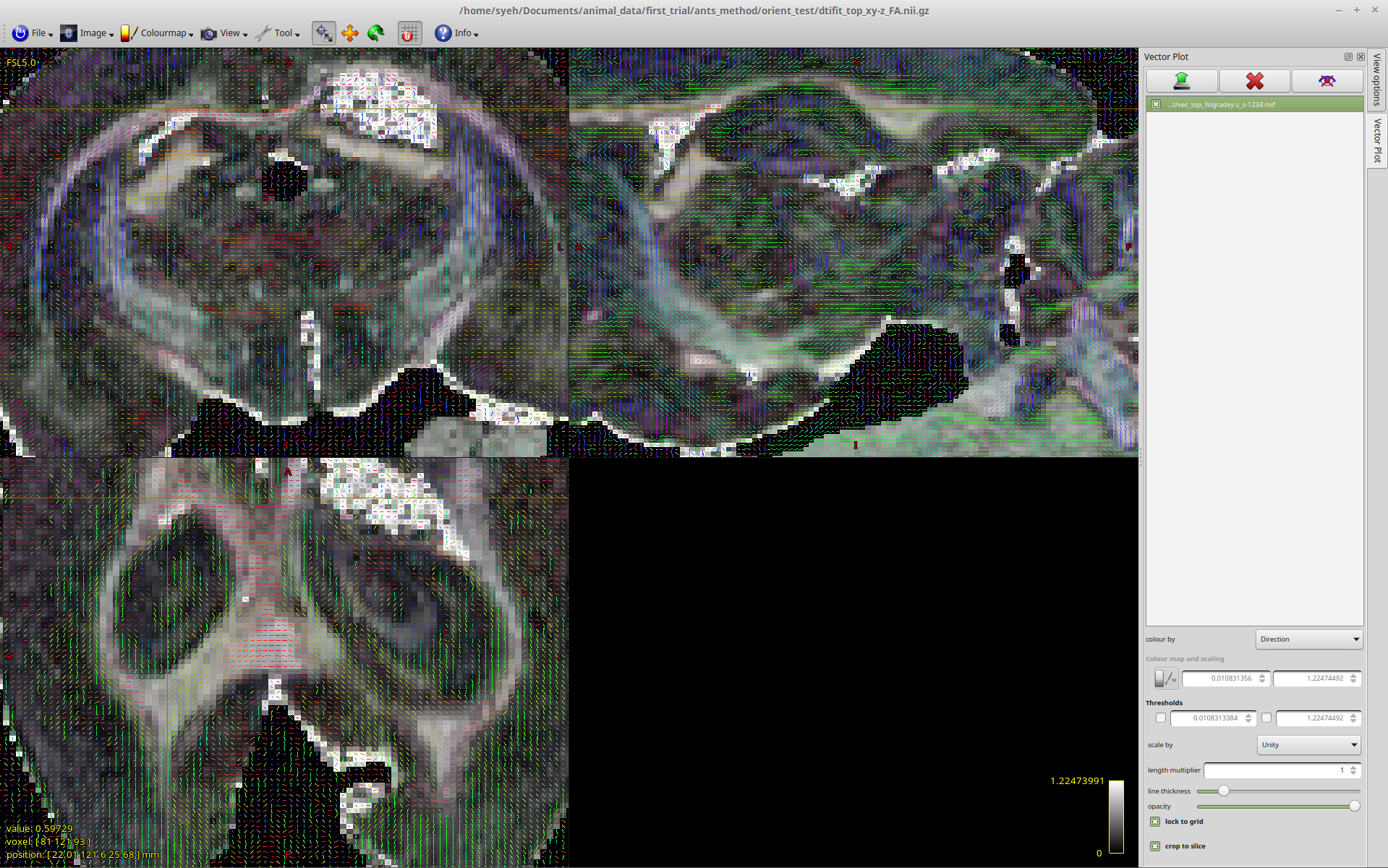
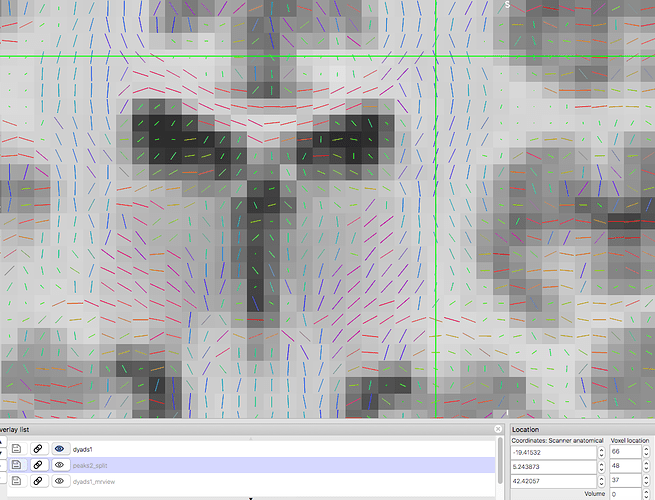
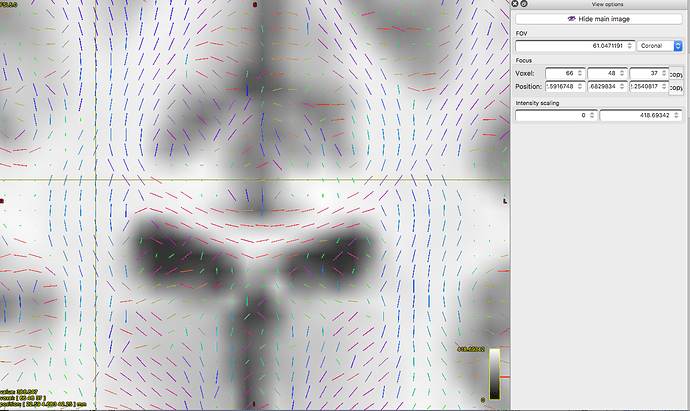
Vectors generated using MRTrix. Note that the previously blue nerve bundle now is green. Vector directions are the same as previous image.This is where the rabbit hole starts.
I have since then played with the strides, mrtransform, manually edited the gradient files using trial and error, and nothing works. It’s especially confusing when you have multiple factors that could all be a contributing factor.
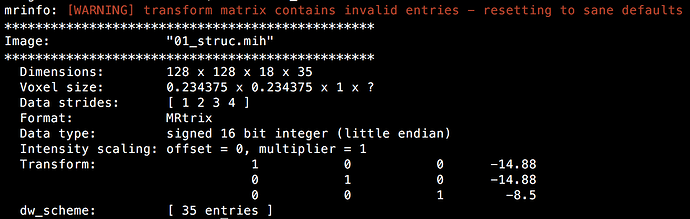
The closest I’ve come to getting anywhere is if I convert the DWI to mif format using mrconvert and then convert back to nifti:
mrconvert -info -stride 1,2,3,4 -fslgrad ../../../bvecs_xy-z.txt ../../../bvals.txt ../dwi_top.nii.gz dwi_top_fslgradxy-z_s1234.mif
mrconvert -info -export_grad_fsl exp_bvecs2.txt exp_bvals2.txt dwi_top_fslgradxy-z_s1234.mif dwi_top_fslgradxy-z_s1234_conv.nii.gz
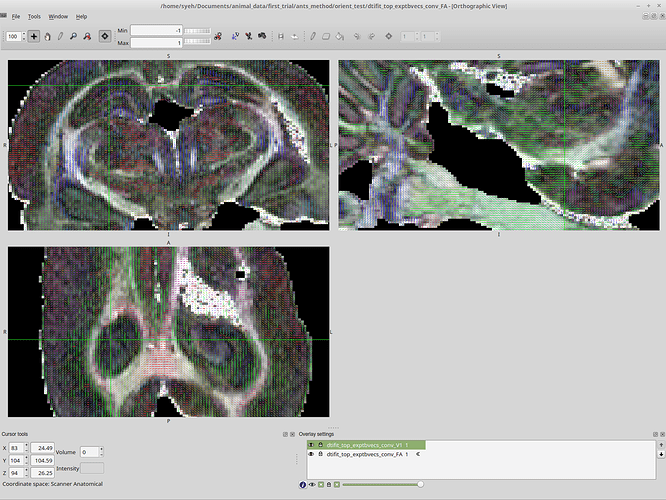
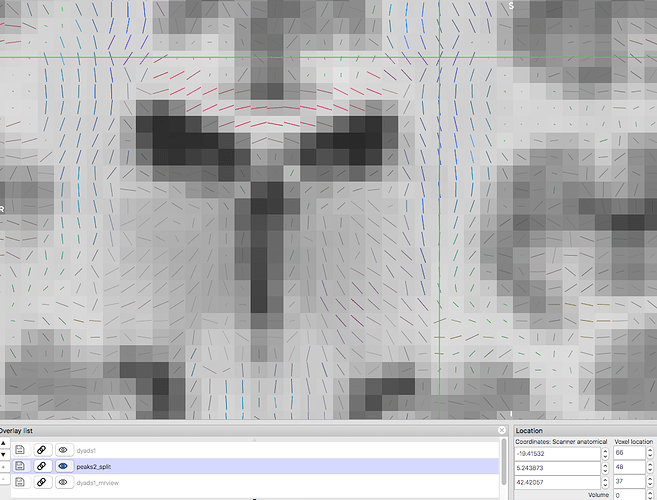
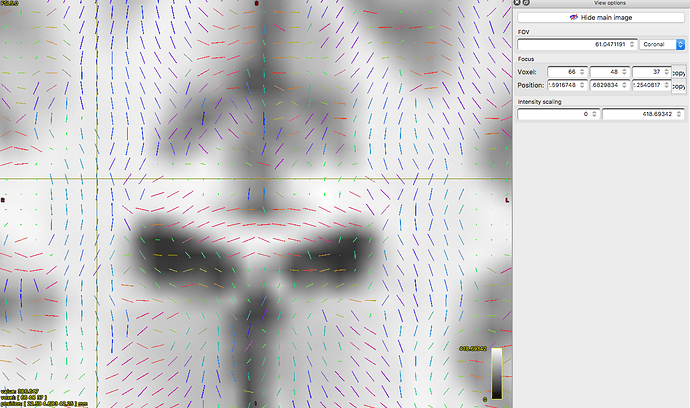
Then, I run FSL’s DTIFIT on the new nifti file, using the exported bvecs and bvals (which are quite different than the original). This is what I get:
This is at least consistent with the MRTrix result, the second image. The vector directions are all the same as the MRTrix result, right down to the green instead of blue nerve bundle.
Where do I go from here? What I want is to display the nerve bundle as blue, which I thought would’ve been a trivial matter. However, it has been anything but trivial.
-Shawn