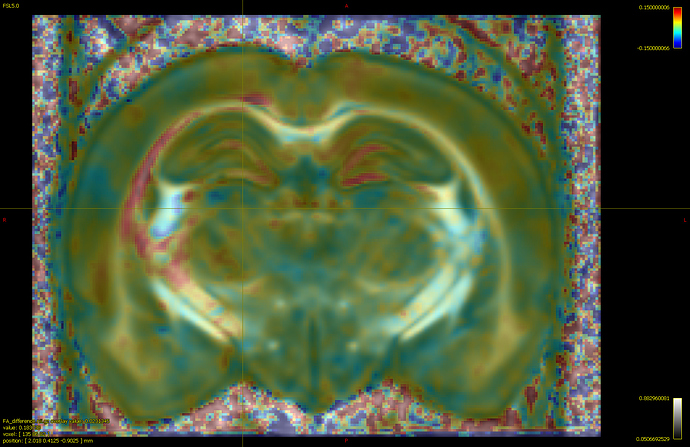
If I load the abs_effect file on the fod template and threshold, color it by an arbitrary value, would it reflect the same?
Is it the same as having subtracted beta1 (control mean) - beta0(tbi mean)?
The absolute effect size is the inner product between the contrast and the beta coefficients (code). So assuming that:
- The first column contains 1s for TBI subjects and 0s for control subjects;
- The second column contains 1s for control subjects and 0s for TBI subjects;
- Your contrast is
[ -1 1 ]
, then yes, this is equivalent to beta1 - beta0. But this is not necessarily true for any arbitrary experimental design.
How would I represent the percentage effect correctly? Is it just loading the percentage effect map and thresholding it according to the fwe values (0.95-1)?
This has to do with being very specific about what data you want to inform what aspect of the visualisation. Typically yes, one would use the percentage effect data to set the colour of each fixel, and the FWE (1 - p-value) image to threshold the fixel display. But it’s best to be explicit about this, rather than “loading the percentage effect map” 
But, when I run it for fdc i get the error: subject data contains non-finite values: -nan.
Given the analysis of FD works without issue, this can only occur if there are non-finite values in your FC somewhere. You could find these using:
mrcalc FC.mif -isnan badFC.mif
, and looking for the fixels that light up in mrview. You could even take the sum of these images across subjects, to find the issue(s) more quickly. Exactly where these are, and how prevalent they are, might provide some insight into what’s going wrong.
Is there a way to override this by setting the fixels which are nan to zero?
You can; but it would be better to first figure out why you are getting NaN values before you go about masking them.
Is there any sense in even trying to extend the fba to include more of the gray matter?
This depends as much on your hypothesis as the underlying techniques used. Generally the fixel correspondence in these areas is poorer, and hence the within-group variances are very large.
With regards to the above post, is it advisable to lower the fmls peak value (fod2fixel) from the default 0.1 in some scenarios?
You certainly can; there’s no “correct” value. Generally for hard-constraint / multi-tissue CSD the FOD amplitudes are smaller than single-tissue soft-constraint CSD, so you can get away with smaller thresholds before you begin to include spurious data. But bear in mind that technically each time you try a different set of masking / segmentation thresholds and re-test your hypothesis, you should be adjusting your p-value threshold for multiple comparisons 
Rob

 (too high threshold tends to cut off the white matter areas and induce holesl!)
(too high threshold tends to cut off the white matter areas and induce holesl!)






 but there’s still 7 hours left… Loads of time!
but there’s still 7 hours left… Loads of time! 

 )
)