Hello,
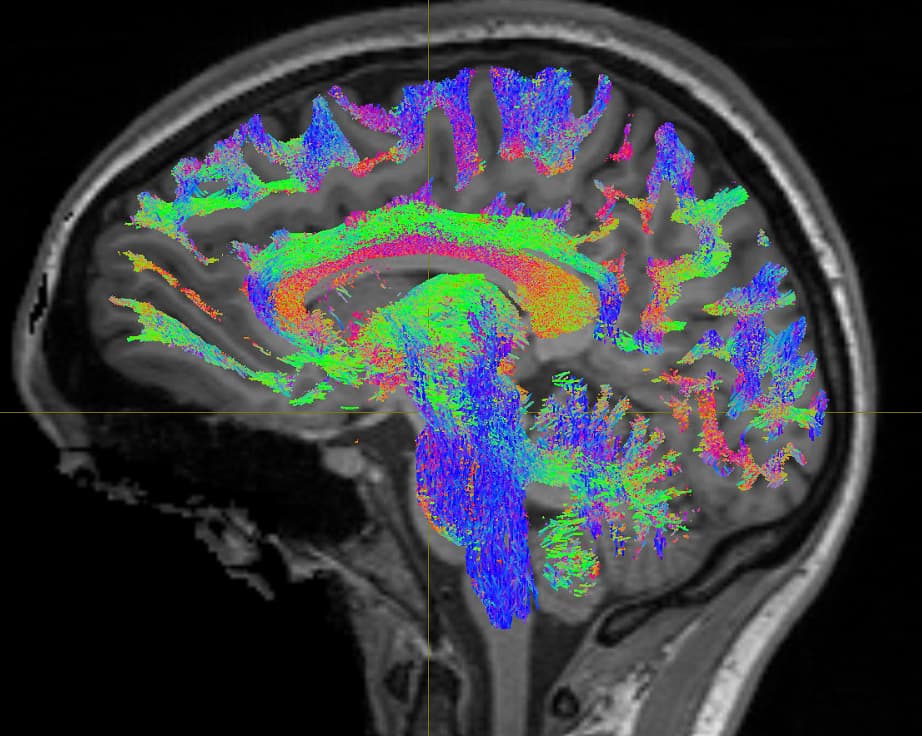
I have created the Whole-brain tractogram using following commands
Step 3.1 - Preparing Anatomical constrained tractography (ACT)
Step 3.1.1 - Preparing Mask for streamline termination
5ttgen fsl T1w.mif 5tt.mif
dwiextract sub_unbiased.mif - -bzero | mrmath - mean mean_b0.mif -axis 3
mrconvert 5tt.mif 5tt.nii.gz
mrconvert mean_b0.mif mean_b0.nii.gz
fslroi 5tt.nii.gz 5tt_gm.nii.gz 0 1
flirt -in mean_b0.nii.gz -ref 5tt_gm.nii.gz -interp nearestneighbour -dof 6 -omat diff2struct_fsl.mat
transformconvert diff2struct_fsl.mat mean_b0.nii.gz 5tt.nii.gz flirt_import diff2struct_mrtrix.txt
mrtransform 5tt.mif -linear diff2struct_mrtrix.txt -inverse 5tt_coreg.mif
Step 3.1.2 - Preparing a mask for streamline seeding
5tt2gmwmi 5tt_coreg.mif gmwmSeed_coreg.mif
Step 3.2 - Creating streamlines
time tckgen -act 5tt_coreg.mif -backtrack -seed_gmwmi gmwmSeed_coreg.mif -maxlength 250 -cutoff 0.06 -select 10000000 wmfod_norm.mif tracks_10M.tck
Step 3.3 - Reducing the number of streamlines
tcksift2 -act 5tt_coreg.mif -out_mu sift_mu.txt -out_coeffs sift_coeffs.txt -nthreads 8 tracks_20M.tck wmfod_norm.mif sift_10M.txt
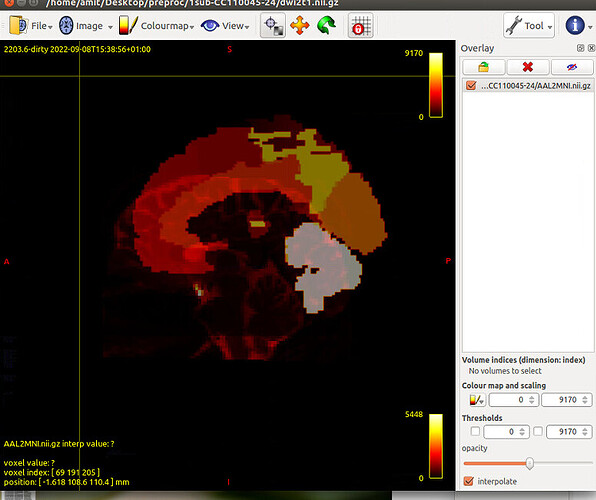
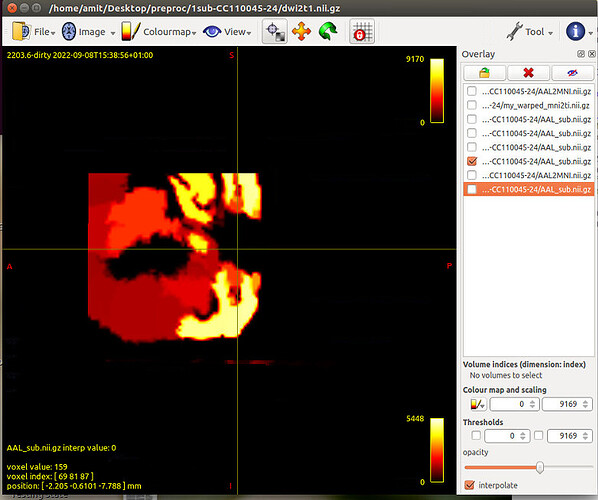
Next, to create AAL percillated file in subject space
bet2 T1w.nii.gz T1_masked.nii -m -f 0.3
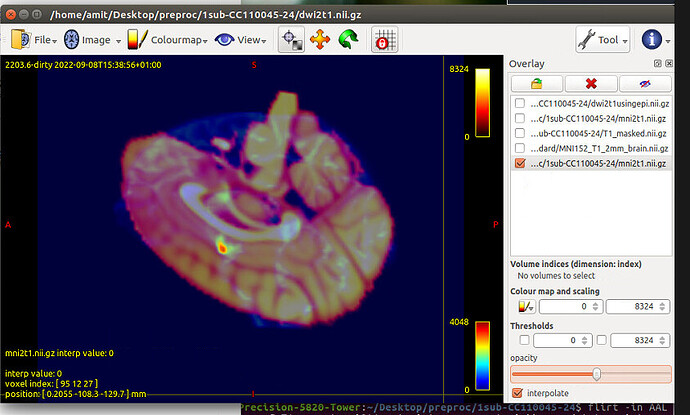
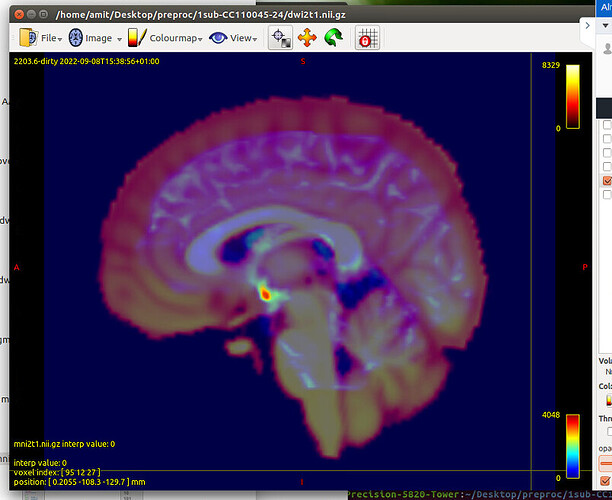
flirt -in T1_masked.nii.gz -ref $FSL_DIR/data/standard/MNI152_T1_1mm_brain.nii.gz -dof 12 -omat T12MNI.mat -interp nearestneighbour -datatype int -out T12MNI.nii.gz
convert_xfm -omat MNI2T1.mat -inverse T12MNI.mat
flirt -in AAL.nii.gz -ref T1_masked.nii.gz -applyxfm -init MNI2T1.mat -interp nearestneighbour -out AAL_sub.nii
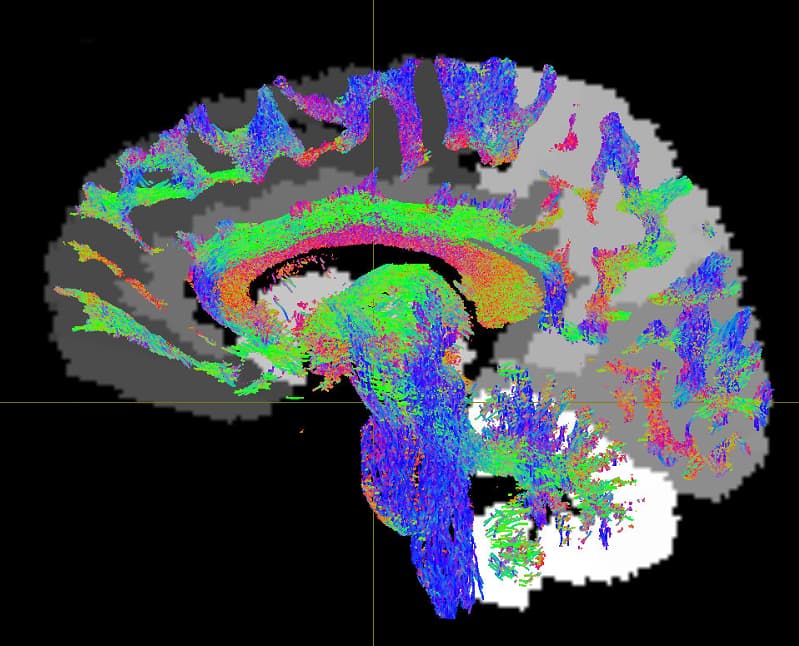
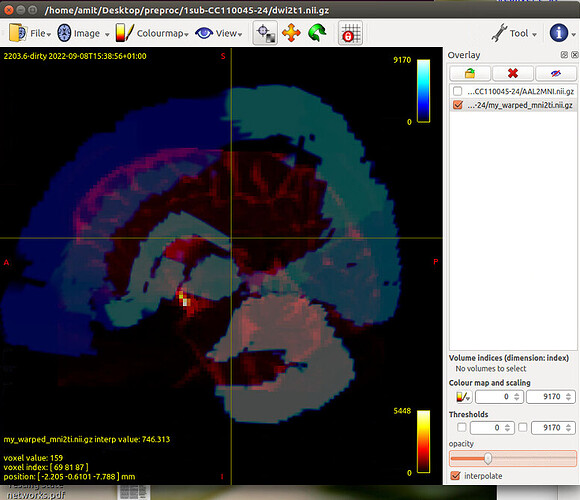
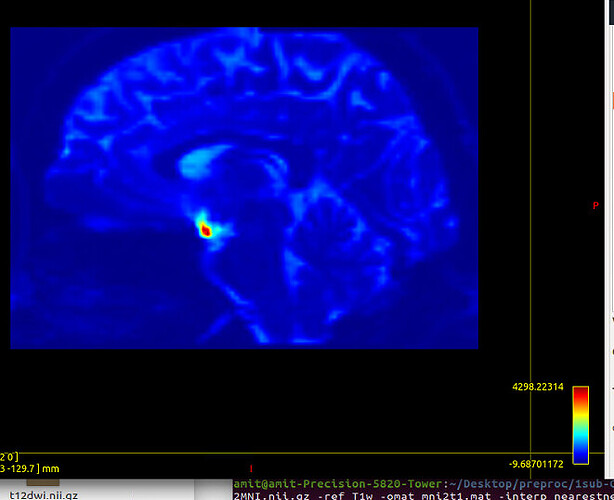
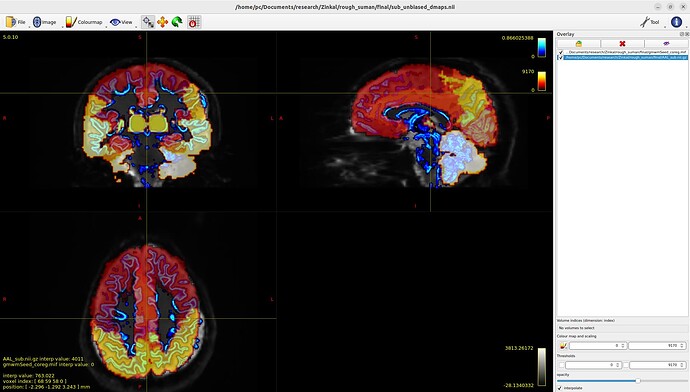
Tract file is not properly register in subject space. This may cause poor registration error while using tck2cinnectome for SC.
tck2connectome: [WARNING] The following nodes are missing from the parcellation image:
tck2connectome: [WARNING] 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, …
Should I tranform the images to DTI space?
I have been trying to solve the problem for two years. Any kind of help would be greatly appreciated.